Massive Eco-Evolutionary Synthesis Simulations (MESS) - CLI Tutorial¶
This is the first part of the full tutorial for the command line interface (CLI) for MESS. In this tutorial we’ll walk through the basics of the process of generating simulations. This is meant as a broad introduction to familiarize users with the general workflow, and some of the parameters and terminology. We will use as a template example the the spider community dataset from La Reunion published by Emerson et al (2017). However, you can follow along with one of the other example datasets if you like, the procedure will be identical although your results will vary.
- Quickstart guide for the impatient
- Equilibrium theory of island biogeography and ecological neutral theory
- Overview of MESS simulation and analysis workflow
- Installation
- Getting started with the MESS CLI
- Create and edit a new parameters file
- Run simulations using your edited params file
- Inspect the output of the simulation runs
- Setting prior ranges on parameters
Each grey cell in this tutorial indicates a command line interaction.
Lines starting with $ indicate a command that should be executed in
a terminal connected to the cluster, for example by copying and pasting
the text into your terminal. Elements in code cells surrounded by angle
brackets (e.g. <mystuffs>) are variables that need to be replaced by
the user. All lines in code cells beginning with ## are comments and should
not be copied and executed. All other lines should be interpreted as output
from the issued commands.
## Example Code Cell.
## Create an empty file in my home directory called `watdo.txt`
$ touch ~/watdo.txt
## Print "wat" to the screen
$ echo "wat"
wat
Quickstart guide for the impatient¶
TL;DR: Just show me how to do the simulations! Say you’re impatient and want to skip right to the good stuff, well here you go.
## Create a parameters file
$ MESS -n simdata
## Do 10 simulations using the default settings and 4 cores
$ MESS -p params-simdata.txt -s 10 -c 4
-------------------------------------------------------------
MESS [v.0.1.1]
Massive Eco-Evolutionary Synthesis Simulations
-------------------------------------------------------------
Project directory exists. Additional simulations will be appended.
<MESS.Region simdata: ['island1']>
establishing parallel connection:
host compute node: [4 cores] on goatzilla
Generating 10 simulation(s).
[####################] 100% Performing Simulations | 0:00:28 |
[####################] 100%
Finished 10 simulations
BAM!
Equilibrium theory of island biogeography and ecological neutral theory¶
Before we begin, if you are unfamiliar with community ecology theory it might be useful to review a brief google presentation given at a recent workshop to provied background and a brief introduction to the MESS model:
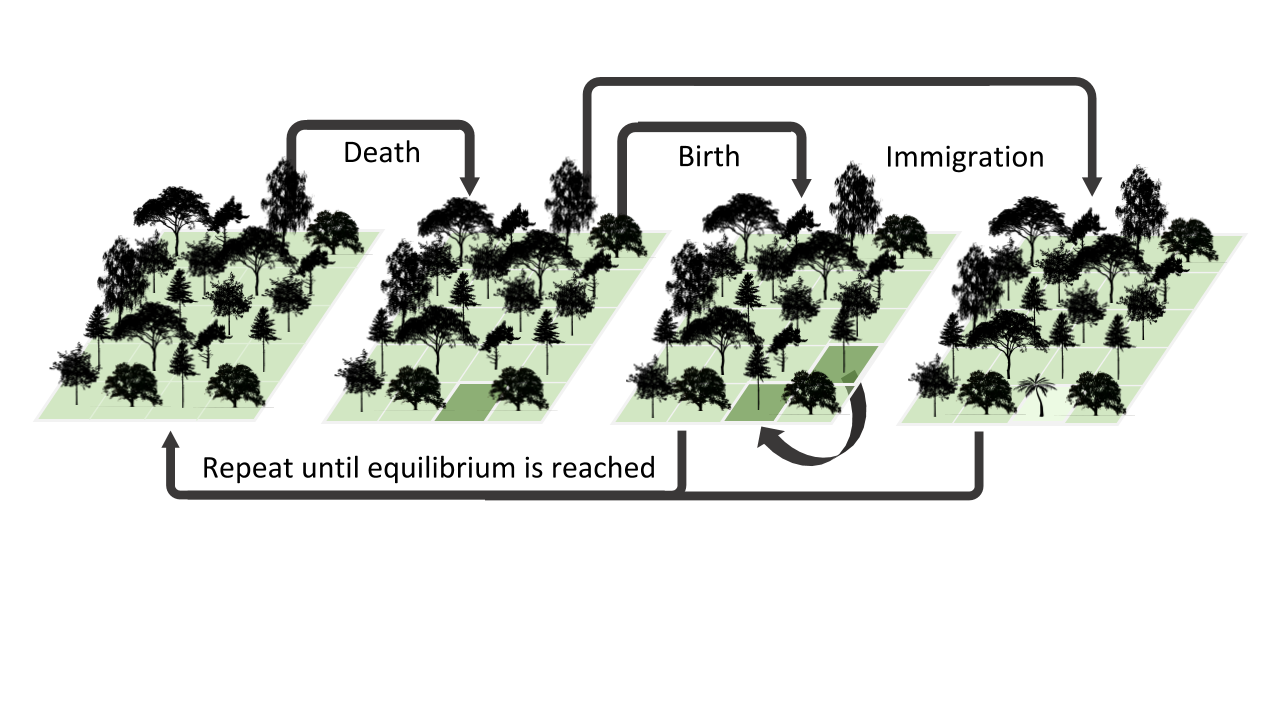
A classic individual based birth/death/colonization model of community assembly. For each timestep one individual is randomly sampled to ‘die’ and is replaced with some small probability by a colonizer from the regional pool, and otherwise is replaced by the offspring of a random individual sampled from the local community.
Overview of MESS simulation and analysis workflow¶
The basic steps of this process are as follows. In this intro tutorial we will only cover steps 1 and 2, but after you are comfortable with generating simulations you may proceed to the ML inference tutorial <inference.md> to learn about the remaining steps:
- Step 1 - Set parameters based on prior knowledge of empirical system
- Step 2 - Run mega simulations
- Step 3 - Use ML to infer community assembly process
- Setp 4 - Use ML to estimate key community assembly parameters
- Step 5 - ???
- Step 6 - Profit!!
Installation¶
MESS is distributed as a conda package so installation is simple and straightforward. If you don’t already have conda and/or MESS installed, please take a moment to install the software.
Getting started with the MESS CLI¶
From here on out we assume you are sitting at a terminal in a conda environment with MESS installed, ok?
To better understand how to use MESS, let’s take a look at the -h argument
to seek ‘help’. We will use some of the MESS command line arguments in this
tutorial (for example: -n, -p, -s, -c). The complete list of
optional arguments and their explanation can be accessed with the -h flag:
$ MESS -h
usage: MESS [-h] [-n new] [-p params] [-s sims] [-c cores] [-r] [-e empirical]
[-f] [-q] [-Q] [-d] [-l] [--ipcluster [ipcluster]] [--fancy-plots]
optional arguments:
-h, --help show this help message and exit
-n new create new file 'params-{new}.txt' in current
directory
-p params path to params file simulations: params-{name}.txt
-s sims Generate specified number of simulations
-c cores number of CPU cores to use (Default=0=All)
-r show status of this simulation run
-e empirical Validate and import empirical data.
-f force overwrite of existing data
-q do not print to stderror or stdout.
-Q do not print anything ever.
-d print lots more info to mess_log.txt.
-l Write out lots of information in one directory per
simulation.
--ipcluster [ipcluster]
connect to ipcluster profile
--fancy-plots Construct fancy plots and animated gifs.
* Example command-line usage:
MESS -n data ## create new file called params-data.txt
MESS -p params-data.txt ## run MESS with settings in params file
MESS -p params-data.txt -f ## run MESS, overwrite existing data.
Create and edit a new parameters file¶
MESS uses a text file to hold all the parameters for a given community assembly
scenario. Start by creating a new parameters file with the -n flag. This
flag requires you to pass in a name for your simulations. In the example we use
simdata but the name can be anything at all. Once you start analysing your
own data you might call your parameters file something more informative, like
the name of your target community and some details on the settings.
$ cd ~
$ mkdir MESS
$ cd MESS
# Create a new params file named 'simdata'
$ MESS -n simdata
This will create a file in the current directory called params-simdata.txt.
The params file lists, on each line, one parameter followed by a ## mark, then
the name of the parameter and then a short description of its purpose. Lets
take a look at it:
$ cat params-simdata.txt
------- MESS params file (v.0.1.1)---------------------------------------------
simdata ## [0] [simulation_name]: The name of this simulation scenario
./default_MESS ## [1] [project_dir]: Where to save files
0 ## [2] [generations]: Duration of simulations. Values/ranges Int for generations, or float [0-1] for lambda.
neutral ## [3] [community_assembly_model]: Model of Community Assembly: neutral, filtering, competition
point_mutation ## [4] [speciation_model]: Type of speciation process: none, point_mutation, protracted, random_fission
2.2e-08 ## [5] [mutation_rate]: Mutation rate scaled per base per generation
2000 ## [6] [alpha]: Abundance/Ne scaling factor
570 ## [7] [sequence_length]: Length in bases of the sequence to simulate
------- Metacommunity params: --------------------------------------------------
100 ## [0] [S_m]: Number of species in the regional pool
750000 ## [1] [J_m]: Total # of individuals in the regional pool
2 ## [2] [speciation_rate]: Speciation rate of metacommunity
0.7 ## [3] [death_proportion]: Proportion of speciation rate to be extinction rate
2 ## [4] [trait_rate_meta]: Trait evolution rate parameter for metacommunity
1 ## [5] [ecological_strength]: Strength of community assembly process on phenotypic change
------- LocalCommunity params: island1------------------------------------------
island1 ## [0] [name]: Local community name
1000 ## [1] [J]: Number of individuals in the local community
0.01 ## [2] [m]: Migration rate into local community
0 ## [3] [speciation_prob]: Probability of speciation per timestep in local community
Note: What’s the difference between a CLI argument and a MESS params file parameter, you may be asking yourself? Well, MESS CLI arguments specify how the simulations are performed (e.g. how many to run, how many cores to use, whether to print debugging information, etc), whereas MESS params file parameters dictate the structure of the simulations to run (e.g. sizes of communities, migration rates, specation rates, etc).
The defaults are all values of moderate size that will generate ‘normal’
looking simulations, and we won’t mess with them for now, but lets
just change a couple parameters to get the hang of it. Why don’t we
change the name parameter of the local community, ‘island1’ is so
generic!. Pick your favorite island and change the name to this. Let’s
also set J (size of the local community in individuals) equal to 500
as this will speed up the simulations (smaller local communities reach
equilibrium faster).
We will use the nano text editor to modify params-simdata.txt
and change this parameter:
$ nano params-simdata.txt
Nano is a command line editor, so you’ll need to use only the arrow keys on the keyboard for navigating around the file. Nano accepts a few special keyboard commands for doing things other than modifying text, and it lists these on the bottom of the frame. After you are done making the changes your file will now have lines that look like this:
La_Reunion ## [0] [name]: Local community name
500 ## [1] [J]: Number of individuals in the local community
Note: For scientific computing, in almost all cases, spaces in variable names and labels should be considered harmful. Notice here how I replace the space in ‘La Reunion’ with an underscore (_) character, this is common practice that you should adopt.
After you change this parameters you may save and exit nano by typing CTRL+o (to write Output), and then CTRL+x (to eXit the program).
Note: TheCTRL+xnotation indicates that you should hold down the control key (which is often styled ‘ctrl’ on the keyboard) and then push ‘x’.
Once we start running the simulations and performing MESS analyses all
the temp files and directories it needs are created in the
project_dir directory and use the prefix specified by the
simulation_name parameter. Because we use the default
(./default_MESS) for the project_dir for this tutorial, all
these intermediate directories will be of the form:
~/MESS/default_MESS/simdata_*, or the analagous name that you used
for your assembly name.
Note on files in the project directory: MESS relies on the integrity of theproject_directoryfor keeping track of various temporary files used by the simulation/analysis process. One result of this is that you can have multiple simulations of the same community assembly scenario using different parameter settings and you don’t have to manage all the files yourself! Another result is that you should not rename or move any of the files or directories inside your project directory, unless you know what you’re doing or you don’t mind if your simulations/analyses break.
Run simulations using your edited params file¶
Here we will start small and generate 10 simulations using 4 cores, just to get
practice running the sims. After this command finishes we’ll have a batch of 10
new summary statistics in our default_MESS directory:
Special Note: In command line mode please be aware to always specify the number of cores with the-cflag. If you do not specify the number of cores, MESS assumes you want only one of them, which will result in painfully slow simulation runs (serial processing).
## -p the params file we wish to use
## -s the number of simulations to perform
## -c the number of cores to allocate <-- Important!
$ MESS -p params-simdata.txt -s 10 -c 4
-------------------------------------------------------------
MESS [v.0.1.1]
Massive Eco-Evolutionary Synthesis Simulations
-------------------------------------------------------------
Project directory exists. Additional simulations will be appended.
<MESS.Region simdata: ['La_Reunion']>
establishing parallel connection:
host compute node: [4 cores] on goatzilla
Generating 10 simulation(s).
[####################] 100% Performing Simulations | 0:00:46 |
[####################] 100%
Finished 10 simulations
Note: You can see here that MESS is intelligently handling all the parallelization work for you. You tell it how many cores to use with the-cflag and it portions out simulations among all the cores as they become available.
Inspect the output of the simulation runs¶
Simulation parameters and summary statistics are written to the
SIMOUT.txt file in the project_dir, which is by defualt created in the
current woring directory as ./default_MESS. You can check the length of
this file:
$ wc -l default_MESS/SIMOUT.txt
11 default_MESS/SIMOUT.txt
# Use `less` to look inside the file. Use `q` to quit less when you are done.
less default_MESS/SIMOUT.txt
NB: Lines in this file are very long, solesswill wrap the text by default. Turn of line wrapping by typing-Sthen pushingenter, which directslessto turn off line-wrapping. Makes it easier to read.
S_m J_m speciation_rate death_proportion trait_rate_meta ecological_strength generations community_assembly_model
100 750000 2.0 0.7 2.0 1.0 0.0 neutral point_mutation 0.0 2000 570.0 500.0 0.01 0.0 189.0 0.696
100 750000 2.0 0.7 2.0 1.0 0.0 neutral point_mutation 0.0 2000 570.0 500.0 0.01 0.0 43.0 0.238
Setting prior ranges on parameters¶
Rather than explicitly specifying MESS parameters, let’s say you’re
interested in actually estimating parameters from the observed data. We can do
this by simulating over a range of values for each parameter of interest, and
then using the MESS inference procedure to estimate these paramters. Let’s say
you would like to estimate the size of the local community (J) and the
migration rate into the local community (m). Edit your params file again
with nano:
nano params-simdata.txt
and change the following two parameter settings. This time we specify a range of values:
1000-2000 ## [1] [J]: Number of individuals in the local community
0.001-0.01 ## [2] [m]: Migration rate into local community
Note: Saving and quitting fromnano:CTRL+othenCTRL+x
Now run some more simulations (by default MESS will append these new simulations to the extant SIMOUT.txt file):
$ MESS -p params-simdata.txt -s 10 -c 4
-------------------------------------------------------------
MESS [v.0.1.1]
Massive Eco-Evolutionary Synthesis Simulations
-------------------------------------------------------------
Project directory exists. Additional simulations will be appended.
<MESS.Region simdata: ['La_Reunion']>
establishing parallel connection:
host compute node: [4 cores] on goatzilla
Generating 10 simulation(s).
[####################] 100% Performing Simulations | 0:00:46 |
[####################] 100%
Finished 10 simulations
Clean up ipcluster <ipyparallel.client.client.Client object at 0x7f15cc3c9090>
Let’s use cut to look at just the columns we’re interested in (J
and m), which are the 13th and 14th columns.
$ cut -f 13,14 default_MESS/SIMOUT.txt
J m
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
500.0 0.01
1118.0 0.00205
1168.0 0.00172
1515.0 0.00323
1061.0 0.0014
1305.0 0.00859
1434.0 0.00881
1397.0 0.00706
1096.0 0.00509
1889.0 0.00112
1699.0 0.00285
And you’ll see that these parameter values are now taking a range, as we specified. Now you are ready to move on to MESS Machine Learning Inference where you will see how we can analyse massive amounts of simulations under varying parameter ranges with a machine learning framework to estimate parameters of the model from real data.